GeneScreen
Rapid identification of mutations in multiple DNA sequence traces
GeneScreen is a program for the rapid processing of multiple DNA sequence traces, in such a way as to facilitate comparison to a reference sequence and efficient identification of mutations. It allows you to specify multiple different amplicons (as when screening a whole gene) and to perform simple, interactive annotation. It even does a pretty good job of deconvoluting heterozygous indels. Mutation data can be exported easily for display on a web page or import into a database such as LOVD.
GeneScreen is an excellent alternative to the Staden package (hard to master) or Mutation Surveyor (expensive), for laboratories routinely involved in mutation hunting.
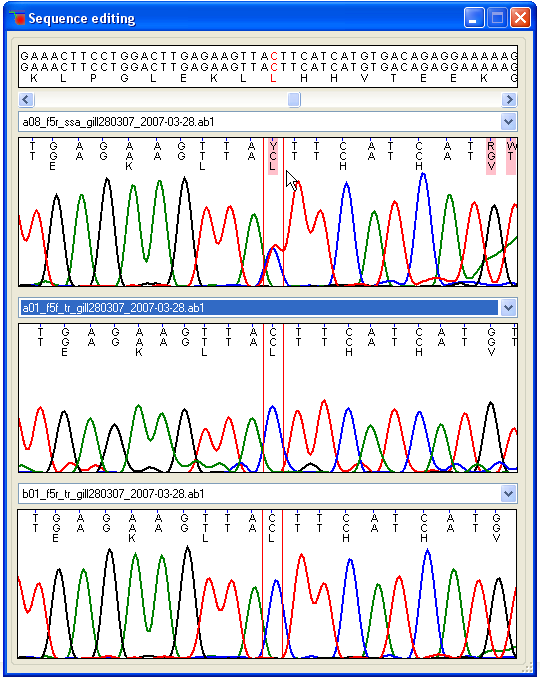
Feature guide
A “walk-through” explanation, with screenshots, of how to use GeneScreen:
HTML guide
(single large page, 950 kB)
GeneScreen reference file creation
The user guide for an application that creates GeneScreen reference files from GenBank files can be found here, while the program can be downloaded from the GeneScreen download page described below
Download
The download page offers the latest version of the GeneScreen program and some sample data files, used in the examples shown in the online Guide.
GeneScreen has been written by Ian Carr. Any feedback on its features or performance would be very welcome.
Citation
Carr IM, Camm N, Taylor GR, Charlton R, Ellard S, Sheridan EG, Markham AF, Bonthron DT.
GeneScreen: a program for high-throughput mutation detection in DNA sequence electropherograms
Journal of Medical Genetics. 2011 Feb;48(2):123-30.