User guide
Introduction
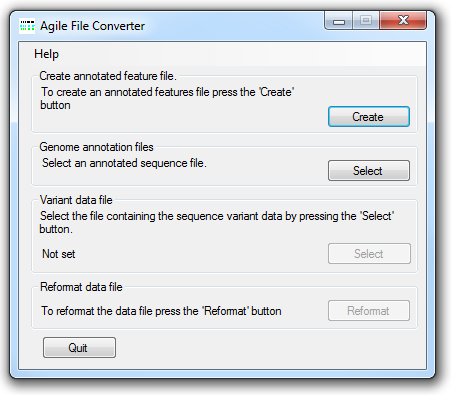
Figure 1: AgileFileConverter's user interface.
AgileFileConverter reformats variant data saved in a tab-delimited text file to a format that the Agile suite of programs can read (see here for a description of this format). As well as reformating the variant file AgileFileConverter also annotates the variants. To achieve this, AgileFileConverter makes use of an annotation file that contains the locations of protein-coding exons and their genomic sequences. This file can be created by AgileFileConverter or by AgileAnnotator. To create an annotation file, first press the button in the Create annotated feature file panel (Figure 1). This causes the Annotation file creation window to be displayed (Figure 2)
Creating an annotation file
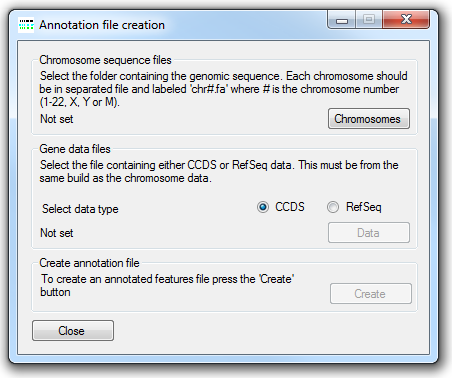
Figure 2: The annotation file creation window.
Note: The genomic sequences in the annotation files are derived from the current genomic sequence reference files and MUST match the genome build used when aligning the sequence reads to the genome and they MUST also be the same build version used by the creators of the CCDS or the RefSeq data files, used to identify the positions of coding sequences. If different reference sequences are used the analysis will fail!
The location of the uncompressed FASTA-format genomic reference files is selected using the button under Chromosome sequence files. These reference sequence files must follow the specific naming convention where each file name starts with "chr" followed by either the chromosome number or "X" or "Y", and has the .fa file extension. Permitted names include chr1.fa, chr5.fa for an autosome, while the X and Y may be named chrX.fa or chr23.fa and chrY.fa or chr24.fa, respectively.
Next select the source of the gene annotation file (CCDS or RefSeq) and then press the button in the CCDS data files panel and select the file containing the positions of the coding sequences as described by the Consensus CDS (CCDS) project or RefSeq datasets. The CCDS file can be downloaded from the NCBI CCDS web page or FTP site. While the RefSeq data file can be obtained from the UCSC Genome Browser's Tables page as described here.
Finally, press the button under Create annotation file and enter a name for the genomic annotation file. Since AgileFileConvertor has to read all of the sequences in the genomic reference files and then write a large amount of data, the creation of the annotation file may take several minutes.
Original file format
The source data file must have the structure shown in Table 1, while the file must contain read depth data, only two of the three read columns shown in the table are needed. For example a valid file could contain the tot_depth and alt_depth, but not the ref_depth columns. The order of the columns in the file is not important however each column must use the header text as shown in the table for AgileFileConvertor to identify each column. If a variant doesn't map to a know variant with a RS ID value the column should contain a '.' character.
| chr_name | chr_start | ref_base | alt_base | tot_depth * | alt_depth * | ref_depth * | dbSNP | gene |
|---|---|---|---|---|---|---|---|---|
| chr1 | 4793 | A | G | 72 | 27 | 45 | rs6682385 | WASH7P |
| chr1 | 7560 | G | C | 43 | 18 | 25 | . | WASH7P |
| chr1 | 7609 | A | G | 18 | 8 | 10 | rs7126006 | WASH7P |
| chr1 | 59374 | A | G | 98 | 96 | 2 | rs2691305 | OR4F5 |
Table 1: While the file must contain read depth data only two of the read depth columns (marked with a * in the table) are required.
Converting a file

Figure 3: The annotation file creation window.
To convert a file, first enter the correct genome annotation file by pressing the on the Genome annotation files panel. Since these files are quite large it may take a few moments to load. Next select the variant tab-delimited variant file by pressing the on the Variant data file. Finally, create the new variant file in the Agile format by pressing the on the Reformat data file panel. The new file will be created with the same name as the original file with '_Agile' appended to the name. If a file with this name already exists, AgileFileConverter will prompt the user to enter a new file name.
Example data
An example file and appropriate annotation file can be downloaded from here (~34 Mb).
Note that this data uses the hg18 genome build co-ordinates.