Walk-through guide
Note: This program requires the Microsoft .NET framework version 2 or later to be installed on your computer.
To annotate variants with regard to the huma genome the Agile suite of programs makes use of an annotation file that contains the locations of protein-coding exons and their genomic sequences. While this file can be created either by other programs e.g. AgileAnnotator or by AgileGenotyperAgileGAFCreator as been developed so that all new updates and improvements can be implemented in one program.
To create an annotation file, first start AgileGAFCreator. This causes the Annotation file creation window to be displayed (Figure 1).
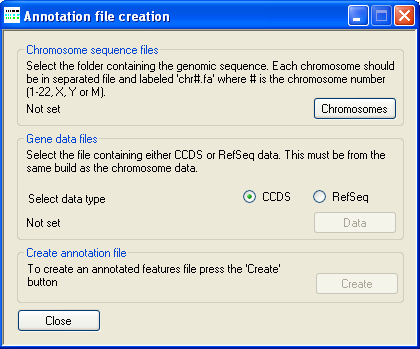
Figure 1: The annotation creation window
Note: The genomic sequences in the annotation files are derived from the current genomic sequence reference files and MUST match the version used when aligning the sequence reads to the genome and they MUST also be the same build version used by the creators of the CCDS or the RefSeq data files, used to identify the positions of coding sequences. If different reference sequences are used the analysis will fail!
The location of the uncompressed FASTA-format genomic reference files is selected using the button under Chromosome sequence files. These reference sequence files must follow the specific naming convention where each file name starts with "chr" followed by either the chromosome number or "X" or "Y", and has the .fa file extension. Permitted names include chr1.fa, chr5.fa for an autosome, while the X and Y may be named chrX.fa or chr23.fa and chrY.fa or chr24.fa, respectively.
Next select the source of the gene annotation file (CCDS or RefSeq) and then press the button in the CCDS data files panel and select the file containing the positions of the coding sequences as described by the Consensus CDS (CCDS) project or RefSeq datasets. The CCDS file can be downloaded from the NCBI CCDS web page or FTP site. While the RefSeq data file can be obtained from the UCSC Genome Browser's Tables page as described here.
Finally, press the button under Create annotation file and enter a name for the genomic annotation file. Since AgileGAFCreator has to read all of the sequences in the genomic reference files and then write a large amount of data, the creation of the annotation file may take several minutes.